Overview of the De Lay Laboratory
What do we study?
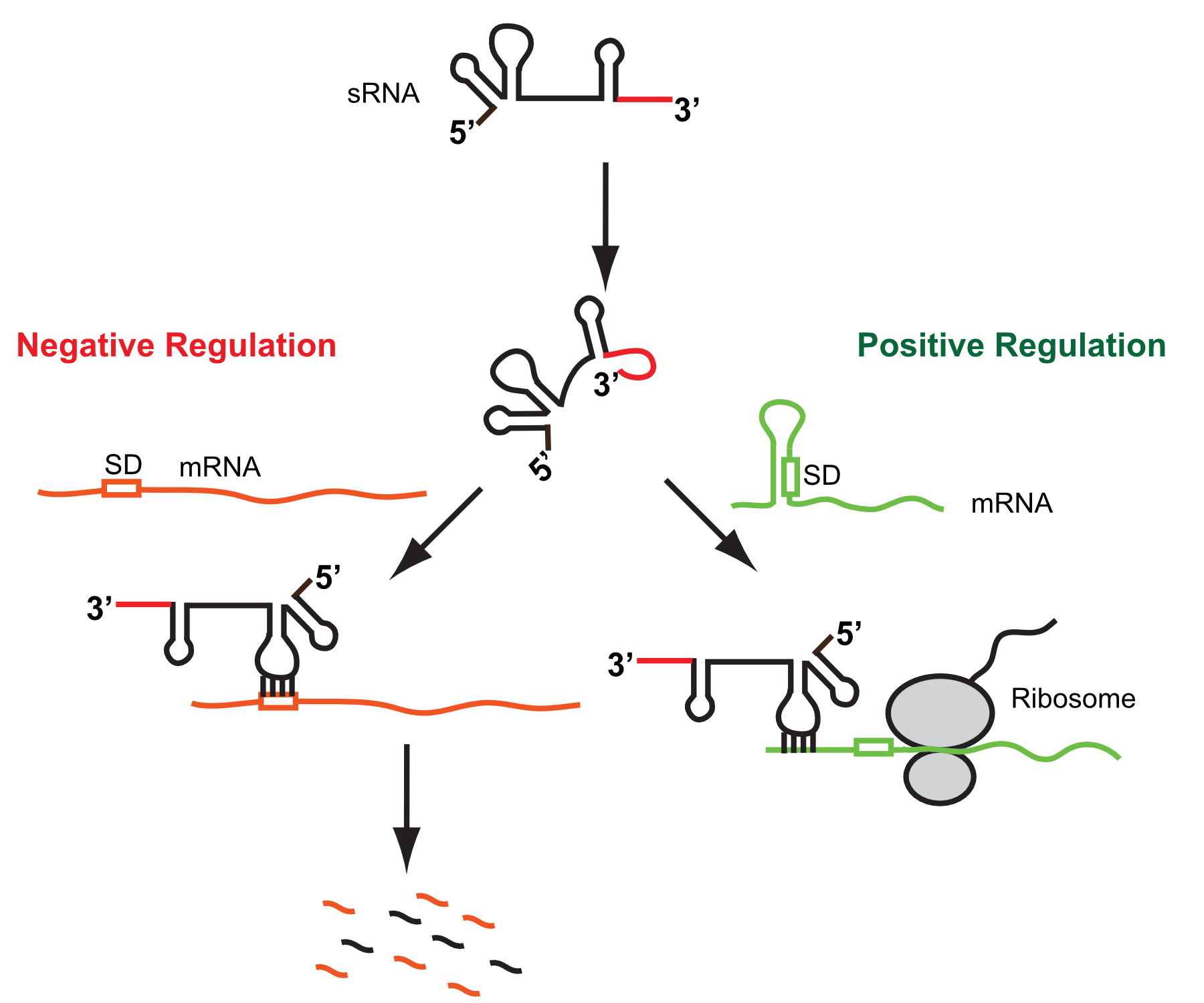
Our research is focused on how small regulatory RNAs (sRNAs) function to regulate gene expression (mainly the production of particular proteins) in the Gram-negative model bacterium Escherichia coli and the Gram-positive bacterial pathogen Streptococcus pneumoniae. Specifically, we are interested in understanding (1) how specific signals lead to production of particular sRNAs, (2) what particular mRNAs are regulated by these sRNAs, (3) how these sRNAs find and bind particular mRNAs in the cell, (4) how binding of these sRNAs to particular mRNAs lead to changes in the production of the encoded proteins, and (5) most importantly, how this alteration in the production of particular proteins impact the physiology and behavior of these bacteria including motility, biofilm development, adherence, metabolism, cell-to-cell communication, antibiotic resistance, and virulence.
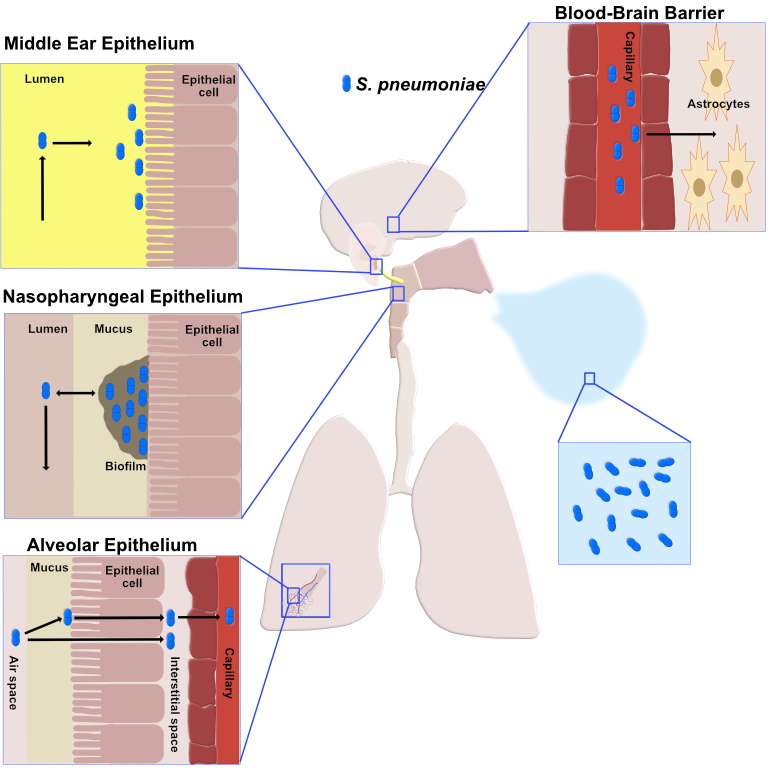
E. coli is a common inhabitant of the human gastrointestinal tract, and while many strains cause no harm or can even be beneficial to humans, others can cause gastroentiritis, diarrhea, bloody diarrhea, neonatal meningitis, and urinary tract infections. S. pneumoniae is common inhabitant of the human upper respiratory tract, where it can reside as a commensal. However, under certain conditions, S. pneumoniae can undergo a change in gene expression causing it to switch to a highly virulent, or disease causing, state. S. pneumoniae can subsequently cause inflammation of the sinuses (sinusitis), then spread into the lungs causing pneumonia, the blood causing sepsis, and/or the brain causing meningitis. S. pneumoniae is a leading cause of bacterial pneumonia, meningitis, and sepsis in the United States and throughout the world.
Why do we study this?
Our overall goal is to understand the biology of E. coli and S. pneumoniae and how these organisms can cause disease in humans. We expect that our studies of sRNAs will help us to better understand how these organisms sense specific signals or stresses, how these signals lead to changes in gene expression, and how these changes in gene expression consequently result in a change in the physiology or behavior of these organisms. For example, studies of a particular sRNA may help us to understand how S. pneumoniae senses a specific signal and how sensing of that signal (1) stimulates S. pneumoniae cells to switch from a human commensal to a hypervirulent pathogen or (2) causes S. pneumoniae cells to spread into the lungs, blood, or brain.
What are small regulatory RNAs (sRNAs)?
RNAs are important biological molecules that perform many roles in bacterial cells from providing the instructions to make the proteins that comprise the cell (mRNAs) to forming the core components of the translational machinery (Ribosomes (rRNA) and tRNAs) that actually synthesize proteins from these instructions. Small regulatory RNAs (sRNAs) are typically 100 nt or less in size (hence small) and function by regulating gene expression at the co-transcriptional or post-transcriptional level, i.e., these sRNAs interact with particular mRNAs and can regulate their synthesis, turnover rate, or ability to interact with the ribosome and be translated to make proteins.
These sRNAs recognize distinct mRNAs through base-pairing between complementary sequences, and a particular sRNA can recognize up to 100s of different mRNAs; base-pairing between the sRNA-mRNA also causes changes, i.e., increases or decreases, in the synthesis, translation, or stability of the mRNA depending on the structure of that mRNA and the precise site of interaction. In general distinct sRNAs are expressed in response to particular signals or stresses. Upon being expressed, sRNAs regulate specific mRNAs to alter the production of particular proteins, which in turn allows the cell to effectively respond to that initial signal or stress. sRNAs have been shown to be key regulators of nearly every aspect of bacterial physiology and behavior including swimming and swarming motility, adherence to biotic and abiotic surfaces, biofilm development, the transport and break down of sugars, amino acids, and other molecules, the synthesis of most cellular molecules, and in the case of bacterial pathogens, virulence (the ability to cause disease).
Why do cells regulate gene expression via small regulatory RNAs (sRNAs)?
- Allows for more rapid changes in gene expression by acting on the mRNA
- Provides additional sequence space beyond the promoter for gene regulation
- Affords multifactorial gene regulation
